About US
Center for High Throughput Sequencing, Core Facility for Protein Research, Key Laboratory of RNA Biology
SpatialDB is supported by CAS Key Technology Talent Program, National Natural Science Foundation of China [31871307, 31801072, 31701122 and 31520103905], National Key Research and Development Project [2018YFA0106901].
Our previous work
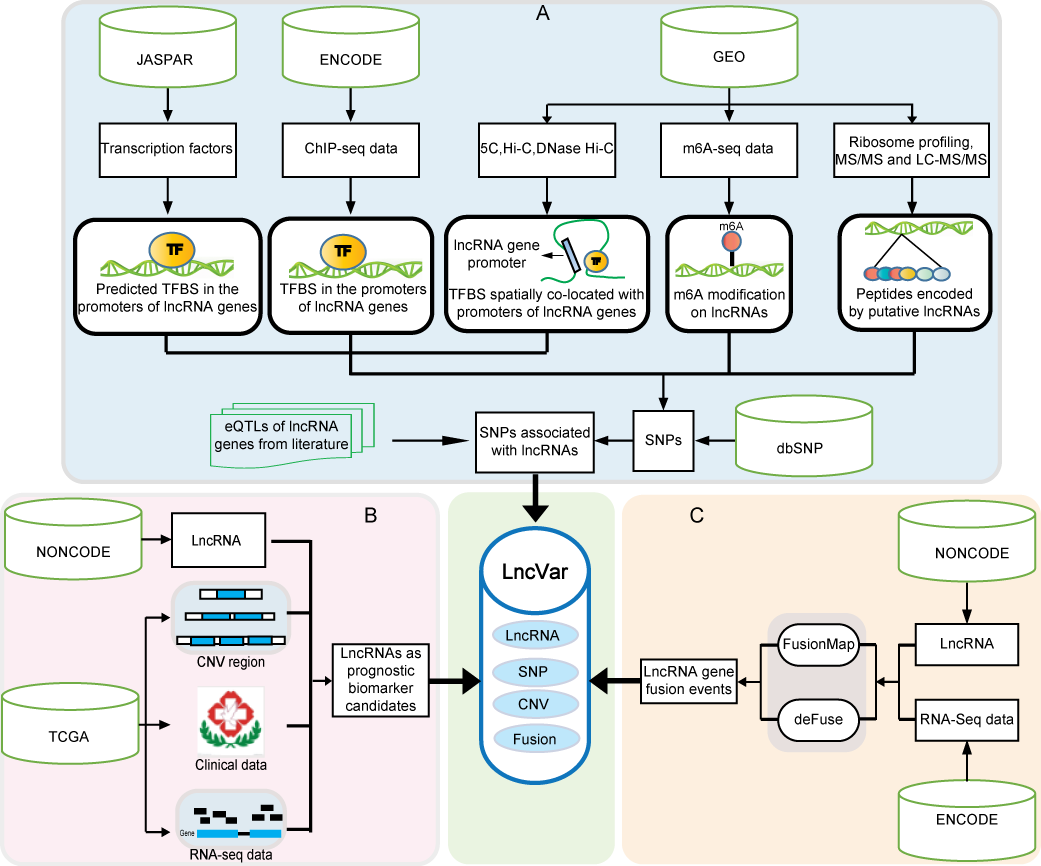
1. LncVar: a database of genetic variation associated with long non-coding genes. (http://159.226.118.31/LncVar/)
Long noncoding RNAs (lncRNAs) have been identified in various species, and play essential roles in many molecular processes. Genetic variations, including single nucleotide polymorphisms (SNPs) and structural variations, are widely distributed in the genome. Variations in the long noncoding gene loci may affect lncRNA transcripts from sequences, structures, expression levels to biological functions.Therefore we constructed LncVar, a database of long noncoding genes associated genetic variations in 6 species (human, mouse, zebrafish, worm, fruitfly, Arabidopsis).
Citation:
1. Chen X., et al. LncVar: a database of genetic variation associated with long non-coding genes. Bioinformatics. 2017 Jan 1;33(1):112-118. doi: 10.1093/bioinformatics/btw581. Epub 2016 Sep 6. PubMed PMID: 27605101.
2. Chen X., et al. LncVar: Deciphering Genetic Variations Associated with Long Noncoding Genes. Methods Mol Biol. 2019;1870:189-198. doi:10.1007/978-1-4939-8808-2_14. PubMed PMID: 30539556.