Dataset details
| Technique | Geo-seq |
|---|---|
| Title | Spatial transcriptome for the molecular annotation of lineage fates and cell identity in mid-gastrula mouse embryo |
| PMID | 27003939 |
| Description | This study documented the spatial transcriptome of the epiblast of the mid-gastrulation (E7.0 late mid-streak stage) mouse embryo, and performed low-input RNA sequencing (RNA-seq) analysis on individual populations (of about 20 epiblast cells each) that were sampled by laser capture microdissection at known positional addresses in single embryos. |
| Source | late mid-streak (∼E7.0) embryos of C57BL/6J mice |
| Species | mouse |
| No. genes | 20,000 genes |
| No. cells | NA |
| Resolution | cellular |
| Data normalization | Z score normalization |
| Data availability | GSE65924 |
Spatially variable (SV) genes
SV genes identified by SpatialDE (Q-value<0.05)
| ID | Gene | Species | Tissue | P-value | Q-value |
|---|---|---|---|---|---|
| ID | Gene | Species | Tissue | P-value | Q-value |
Functional enrichment analysis of SV genes
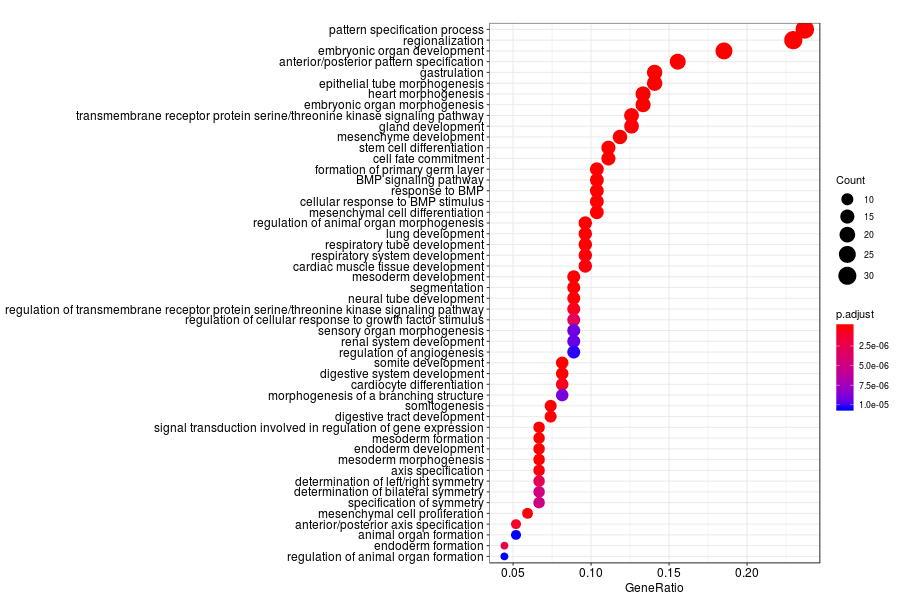
GO enrichment analysis of SV genes identified by SpatialDE (Q-value<0.05) in mid-gastrula mouse embryo.
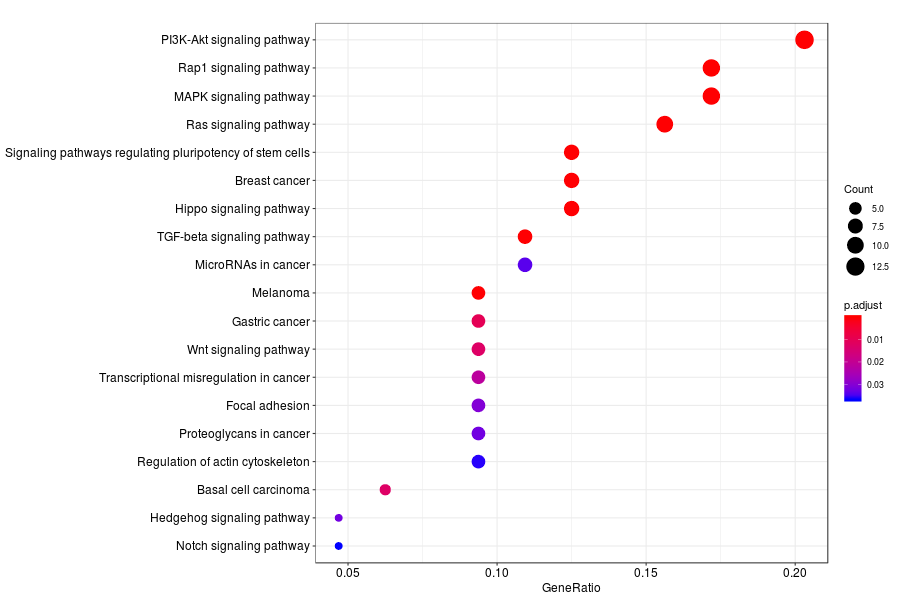
KEGG enrichment analysis of SV genes identified by SpatialDE (Q-value<0.05) in mid-gastrula mouse embryo.