Dataset details
| Technique | Spatial transcriptomics |
|---|---|
| Title | Spatially resolved transcriptomics enables dissection of genetic heterogeneity in stage III cutaneous malignant melanoma |
| PMID | 30154148 |
| Description | Spatial transcriptomics (ST) was adapted to melanoma lymph node biopsies and successfully sequenced the transcriptomes of over 2,200 tissue domains. |
| Source | lymph node metastases from four patients diagnosed with stage III melanoma |
| Species | human |
| No. genes | nearly 18,000 genes |
| No. cells | NA |
| Resolution | 100 um |
| Data normalization | Median ratio normalization |
| Data availability | Count matrixes for exploring genes / gene signatures of interest are available at www.spatialtranscriptomicsresearch.org |
Spatially variable (SV) genes
SV genes identified by SpatialDE (Q-value<0.05)
| ID | Gene | Species | Sample | Array | P-value | Q-value |
|---|---|---|---|---|---|---|
| ID | Gene | Species | Sample | Array | P-value | Q-value |
SV genes identified by trendsceek (Q-value<0.05)
| ID | Gene | Species | Sample | Array | Q-value(Emark) | Q-value(Vmark) | Q-value(MarkCorr) | Q-value(MarkVario) |
|---|---|---|---|---|---|---|---|---|
| ID | Gene | Species | Sample | Array | Q-value(Emark) | Q-value(Vmark) | Q-value(MarkCorr) | Q-value(MarkVario) |
Functional enrichment analysis of SV genes
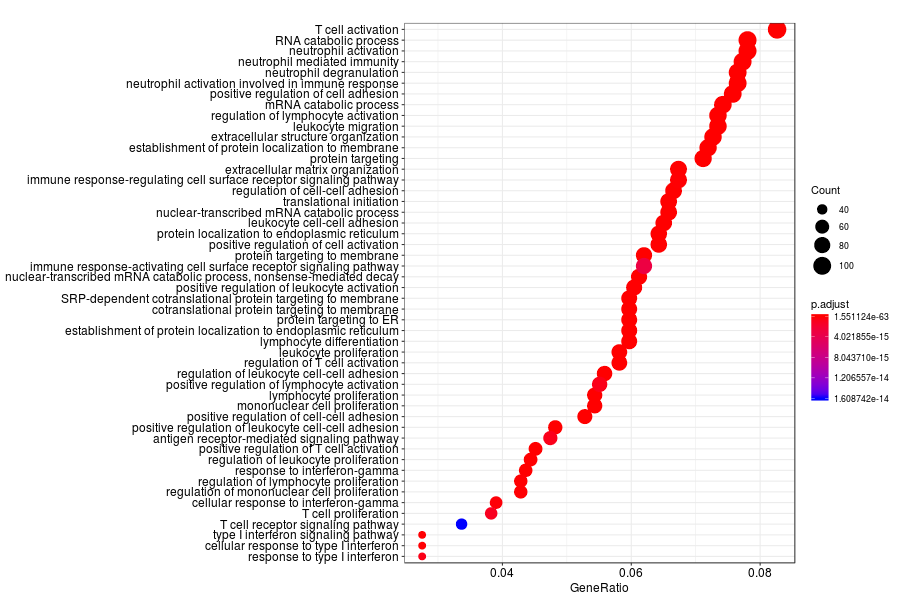
GO enrichment analysis of SV genes identified by SpatialDE (Q-value<0.05) in lymph node metastases from melanoma.
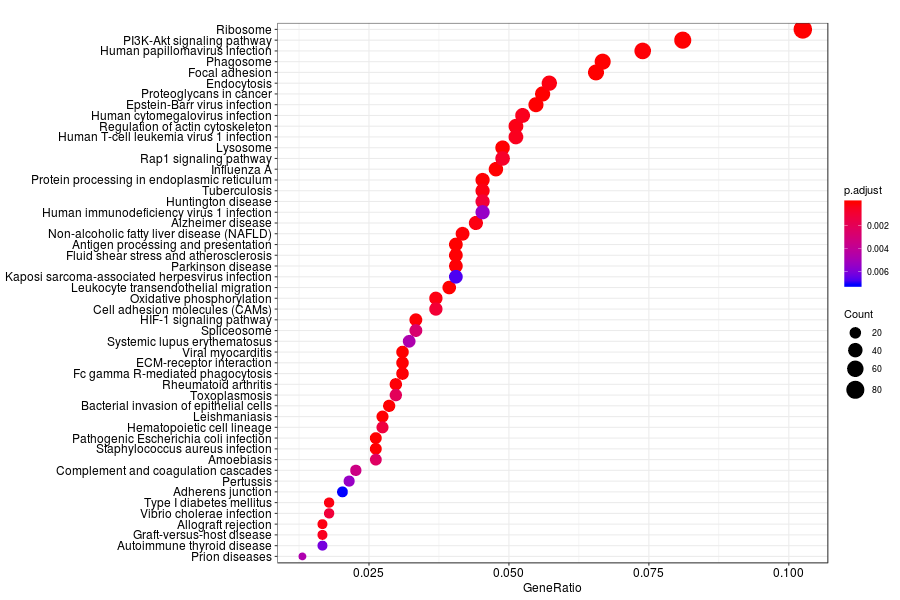
KEGG enrichment analysis of SV genes identified by SpatialDE (Q-value<0.05) in lymph node metastases from melanoma.
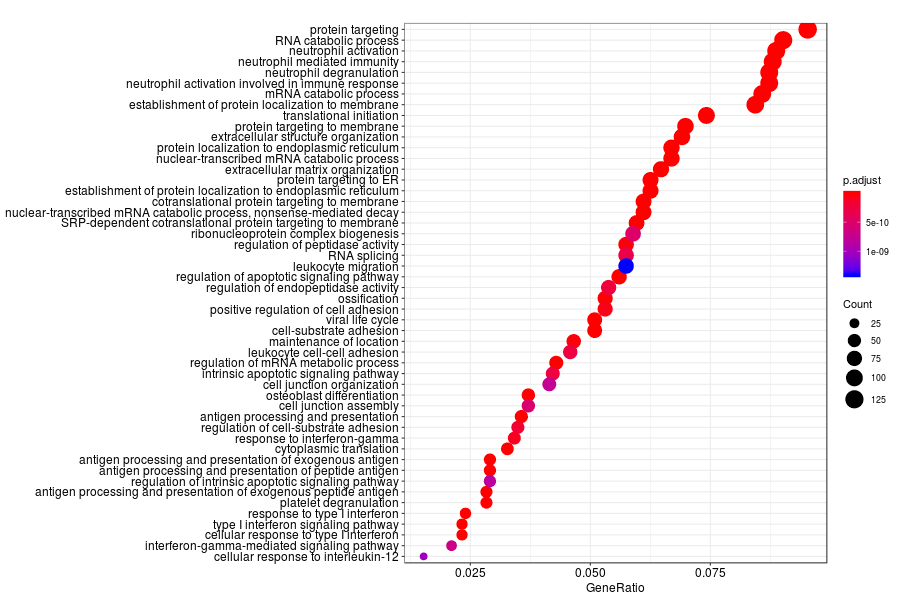
GO enrichment analysis of SV genes identified by trendsceek (Q-value<0.05) in lymph node metastases from melanoma.
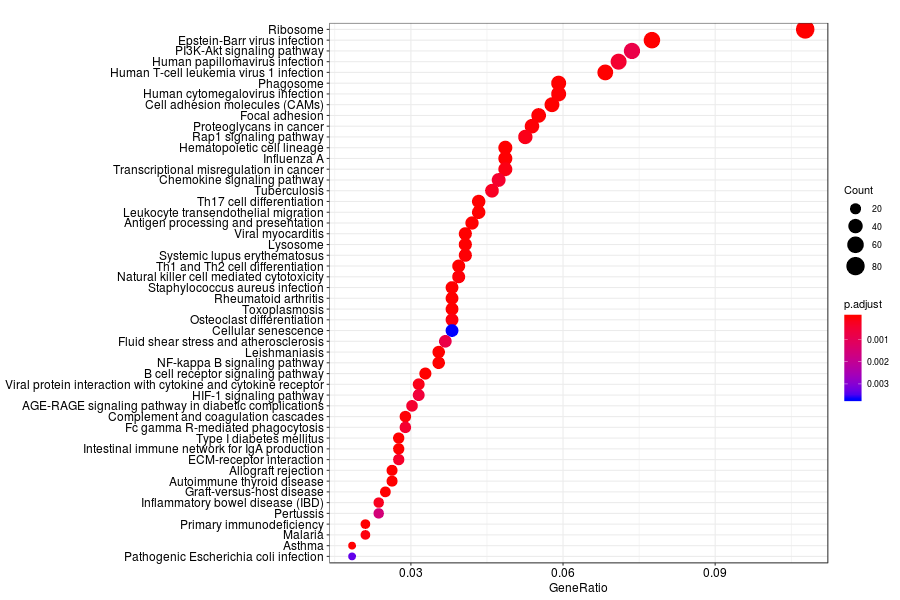
KEGG enrichment analysis of SV genes identified by trendsceek (Q-value<0.05) in lymph node metastases from melanoma.