Dataset details
| Technique | LCM-seq |
|---|---|
| Title | Topographical transcriptome mapping of the mouse medial ganglionic eminence by spatially resolved RNA-seq |
| PMID | 25344199 |
| Description | This study adapted the STRT (single-cell tagged reverse transcription) method to laser microdissected tissue samples. By systematically sampling the mouse medial ganglionic eminence (MGE) tissues in a regular grid, they isolated 50×50×50 μm3 cubes that are akin to the `voxels’ in a three-dimensional volumetric space. |
| Source | wild-type and Gfra1Tlz mutant embryos (both C57bl6/J) |
| Species | mouse |
| No. genes | 14942 genes |
| No. cells | NA |
| Resolution | 50 um |
| Data normalization | TPM |
| Data availability | GSE60402 |
Spatially variable (SV) genes
SV genes identified by SpatialDE (Q-value<0.05)
| ID | Gene | Species | Tissue | Genotype | P-value | Q-value |
|---|---|---|---|---|---|---|
| ID | Gene | Species | Tissue | Genotype | P-value | Q-value |
Functional enrichment analysis of SV genes
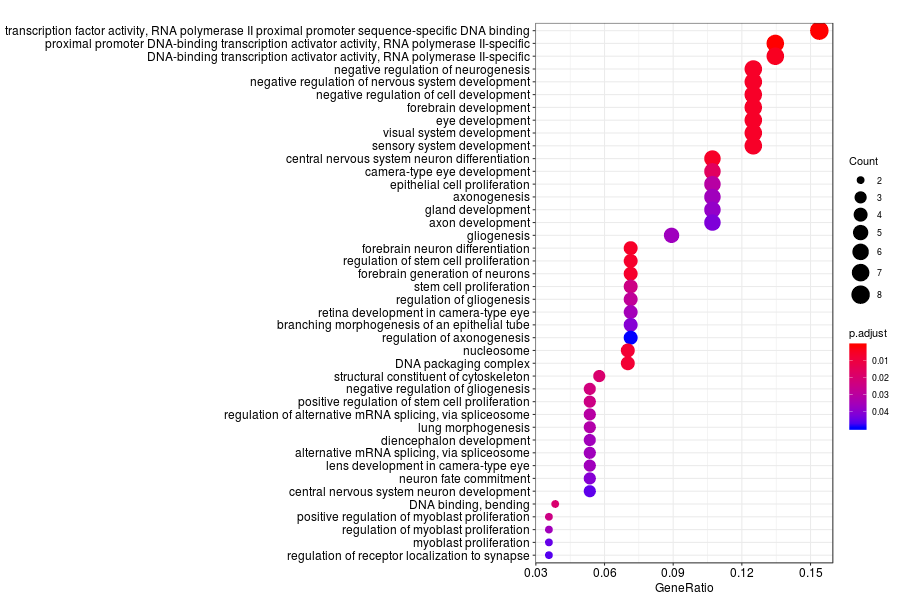
GO enrichment analysis of SV genes identified by SpatialDE (Q-value<0.05) in medial ganglionic eminence of wildtype mouese.
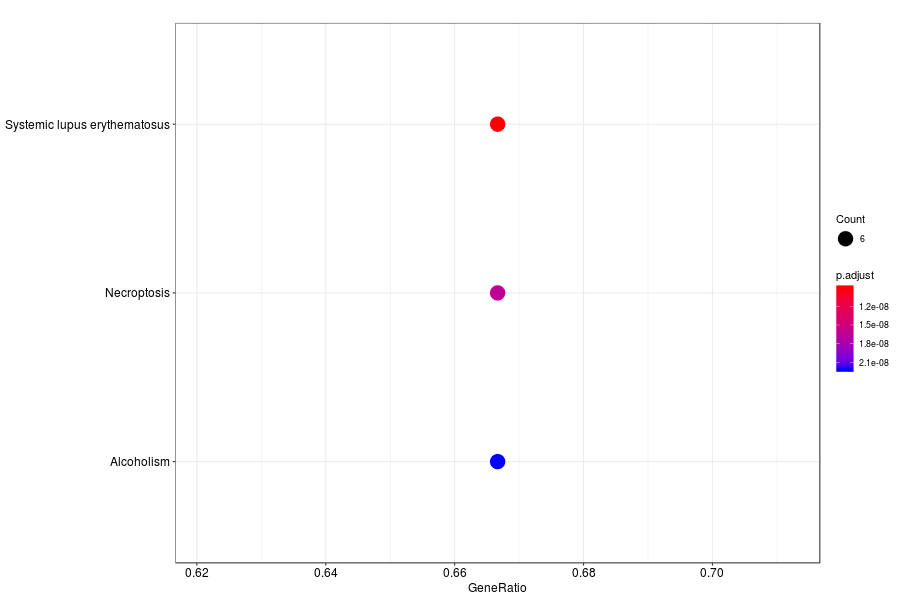
KEGG enrichment analysis of SV genes identified by SpatialDE (Q-value<0.05) in medial ganglionic eminence of wildtype mouese.
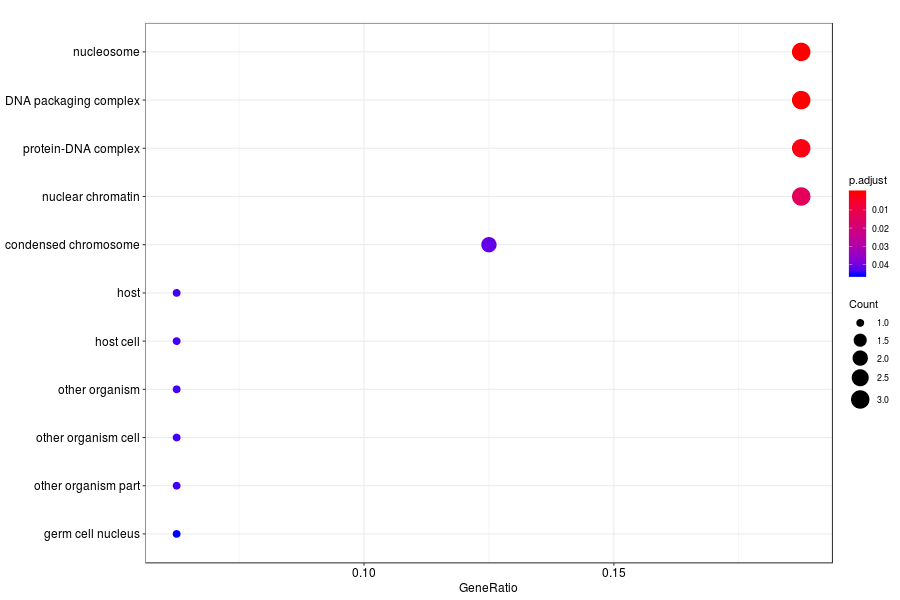
GO enrichment analysis of SV genes identified by SpatialDE (Q-value<0.05) in medial ganglionic eminence of mutation mouese.
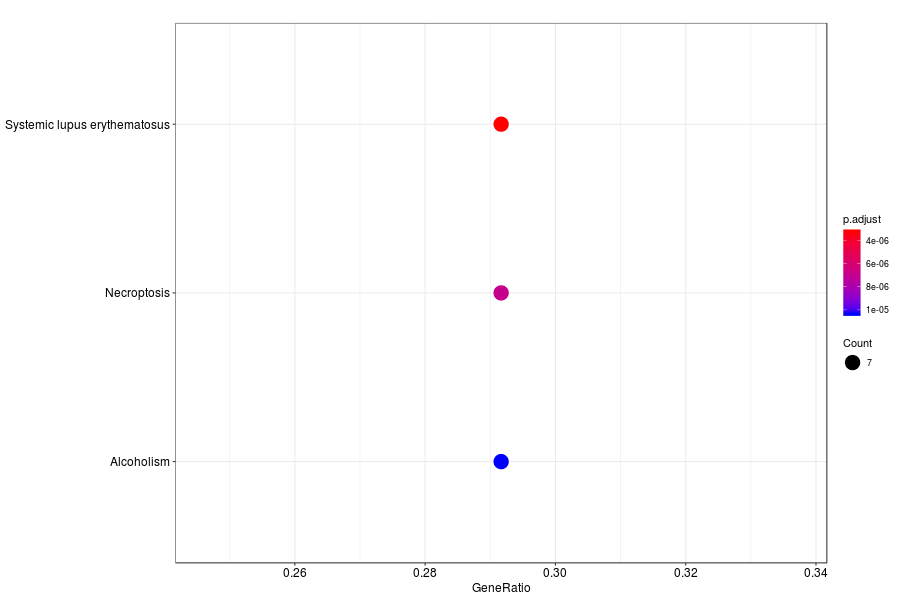
KEGG enrichment analysis of SV genes identified by SpatialDE (Q-value<0.05) in medial ganglionic eminence of mutation mouese.